RESEARCH PROJECTS
The human genome is estimated to contain ~20-25,000 genes, with 50%-75% of multi-exon genes predicting to undergo splicing, considerably amplifying proteomic diversity. Our RTG will investigate how that diversity can be exponentially amplified by studying how certain proteins have multiple functions depending on their localization. Thus, projects in our RTG follow one overarching question: how does dynamic protein (re)localization contribute to cellular adaptation through expanding proteomic complexity and plasticity?
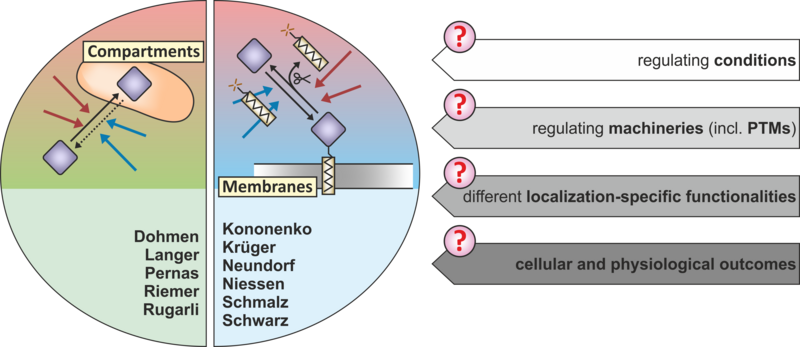
Projects in our consortium fall roughly into two conceptual groups:
- Projects investigating proteins (re)localizing to and from different (sub)compartments. Different compartments/substructures provide different biochemical conditions and interaction partners, that can impact protein function and cellular outcomes. Furthermore, the role of different PTMs such as phosphorylation, proteolytic processing, sumoylation, or thiol modifications in driving protein (re)localization will be addressed.
- Projects investigating proteins (re)localizing to and from membranes. Movement restrictions, integration into signaling pathways, and the hydrophobic interaction and reaction opportunities at membranes will play important roles in elucidating mechanisms driving protein (re)localization between membranes. The contribution of different PTMs will also be assessed in these projects.
